Team : Mucins, Cancer and Drug Resistance
Our team “Mucins, Cancer and Drug Resistance” is composed of researchers, professors, associate professors, clinicians, biologists, pathologists, engineers, technicians and students.
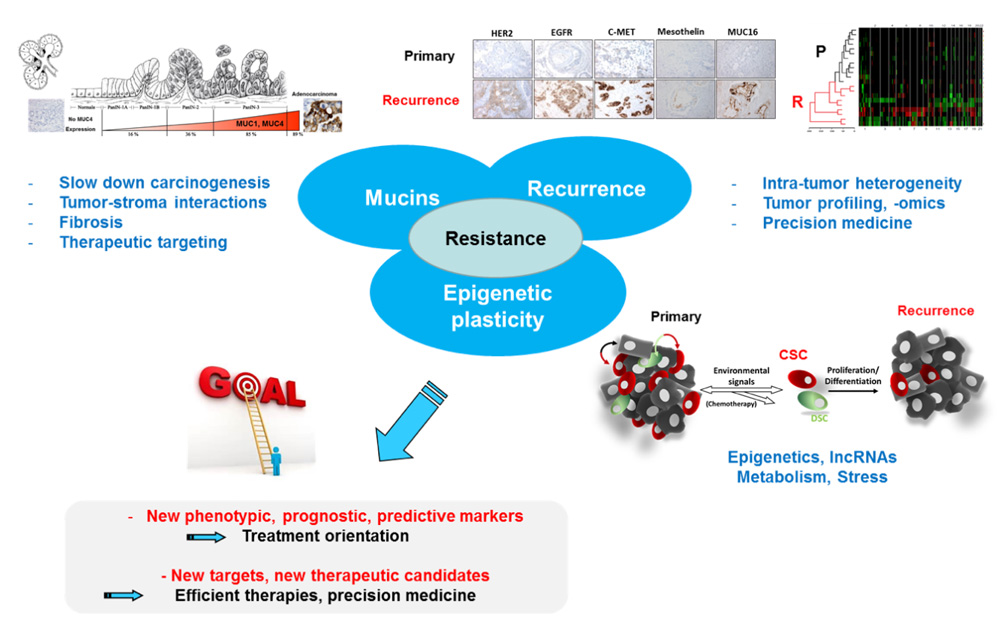
Our research is focused on the role of mucins in cancer for which we are internationally recognized and on the mechanisms of tumor resistance to the chemotherapeutic treatments. Since solid tumors are characterized by an important intra-tumor and micro-environment (presence of different matrix compositions and various cell types: stromal, immune, inflammatory cells…) heterogeneity, we work at better understanding and characterizing (i) the molecular mechanisms involved in chemoresistance (intracellular signaling, epigenetics, metabolism, ncRNAs, regulatory mechanisms…), (ii) the role of tumor-stroma interactions in tumor behavior as well as (iii) the tumor heterogeneity (cartography and molecular profiling, multi-omics and single cell approaches).
For that, we develop a fundamental, pre-clinical and translational research in cancerology in collaboration with hospital departments of the Lille hospital (CHU Lille) with the aim to transfer our data to the clinic (new markers, new therapeutic targets, new protocols, precision medicine) thanks to annotated in-house patient cohorts and national clinical-biological databases.
We also develop interdisciplinary research with chemists (new drugs, therapeutic chemistry), physicists (4D microfluidic models, organ/tumor-on-chip models) and bioinformaticians (Artificial Intelligence for Cancer).
Our long term goals are to discover new phenotypic, prognostic, predictive, treatment response markers (treatment orientation) in oeso-gastric, colonic, pancreatic and renal cancers but also to identify new therapeutic targets (increase treatment efficacy) in order to offer new tools to the clinicians for a better patient management and better patient care (precision medicine).
Every year we host 10-15 students/fellows (basic scientists, MD’s, PharmD’s) for their Master internship, PhD thesis, or post-doctoral fellowship. We are affiliated to the Lille Biology-Health Doctoral School (EDBSL) (http://edbsl.univ-lille2.fr).
We are a major team in cancer research in Lille and we are supported by Inserm (https://www.inserm.fr/), CNRS (http://www.cnrs.fr), Hauts de France region (https://www.hautsdefrance.fr/), ONCOLille institute (https://oncolille.univ-lille.fr/), North-West Canceropole (https://www.canceropole-nordouest.org/), University of Lille (https://www.univ-lille.fr/), the Lille Hospital Center (CHU) (https://www.chu-lille.fr/) and the Regional Center against Cancer Oscar Lambret (COL) (https://www.centreoscarlambret.fr/).
Our research themes are:
A- Role of MUC1 and MUC4 mucins in epithelial carcinogenesis and chemoresistance
A.1- Role of MUC1 and MUC4 mucins in pancreatic, oeso-gastric, colonic and renal carcinogenesis. Study of the molecular mechanisms in in vitro (2D, 3D, organoids and cellular models and 4D microfluidic devices (collaboration with physicists)), in vivo (transgenic mice, cellular and Patient-Derived Xenografts (PDX)) and ex vivo (patient samples, cohorts) models.
A.2- Study of the structure-function relationship of MUC4-ErbB2 complex and of MUC1-CT cleavage site. Therapeutic targeting and discovery of new anti-cancer drug candidates (collaboration with chemists).
B- Study of molecular mechanisms responsible for tumor resistance/recurrence
B.1- Deciphering of resistance mechanisms to chemotherapy and drug escape (tumor dormancy, cellular reprogramming). Epigenetic plasticity of cancer cells (DNA methylation, histone code, noncoding RNA, metabolism, stemness).
B.2- Transversal/translational research with hospital partners (cohorts, CBD): Search for prognostic (identification and classification of tumors) and predictive (therapeutic orientation, patient management) markers/factors. Comparison of primary tumor/recurrence/metastasis. Tumor heterogeneity (molecular cartography of tumors using multi-omics & single cell approaches). Use of Artificial Intelligence (Machine/Deep Learning, collaboration with bioinformaticians).
> SCIENTISTS
> ENGINEERS -TECHNICIANS
> STUDENTS
 | Dr Isabelle VAN SEUNINGEN, chercheur DR1 CNRS, PhD |
 | Dr Vincent Senez, DR1 CNRS, PhD Research: Keywords: microfluidics, micromechanics, electric sensor, tumor-on-chip Collective responsibilities: vincent.senez(@)univ-lille.fr |
 | Dr Nicolas JONCKHEERE, chercheur CRCN Inserm, PhD |
 | Dr Bernadette NEVE, chercheur CRCN Inserm, PhD |
 | Dr Audrey VINCENT, chercheur CRCN Inserm, PhD Responsabilité collective : Responsable de la plate-forme de culture cellulaire 3D OrgaRES, ORGALille et OrgaNO |
 | Dr Michaël PERRAIS, Maître de Conférences MCU, PhD |
 | Pr Emmanuelle LETEURTRE, anatomopathologiste, MD PhD |
 | Pr Marie-Pierre BUISINE, biologiste, PharmD PhD |
 | Pr Guillaume PIESSEN, chirurgien digestif, MD PhD |
 | Dr Florence RENAUD, anatomopathologiste, MD PhD |
 | Pr Sébastien AUBERT, anatomopathologiste, MD PhD |
 | Dr Viviane GNEMMI, anatomopathologiste, MD PhD |
 | Dr Julie LECLERC, biologiste, PharmD PhD |
 | Dr Lucie COPPIN, biologiste, PharmD PhD |
 | Dr Anne-Frédérique DESSEIN, biologiste, PharmD PhD |
 | Pr Stéphanie TRUANT, chirurgien digestif, MD PhD |
 | Pr Pascal PIGNY, biologiste, PharmD PhD |
 | Pr Mohammed HEBBAR, oncologue, MD PhD mohamed.hebbar(@)chru-lille.fr |
 | Dr Clarisse EVENO, Chirurgien, MD PhD |
 | Dr Mehdi EL AMRANI, Chirurgien, MD PhD |
 | Dr Jean-Baptiste GIBIER, anatomopathologiste, MD PhD Recherche : Rôle de MUC1 au cours de l’insuffisance rénale aiguë et chronique. Étude de MUC1 dans la réponse inflammatoire après stress rénal. Étude des mécanismes conduisant à la fibrose rénale. jeanbaptiste.gibier(@)chru-lille.fr |
 | Belinda DUCHÊNE, technicienne de Recherche Inserm belinda.duchene(@)inserm.fr |
 | Marie-Paule DUCOUROUBLE, technicienne de Recherche Inserm marie-paule.ducourouble(@)inserm.fr |
Soumaya EL MOGHRABI, IE Inserm Compétences : biologie cellulaire et moléculaire, modèles cellulaires 3D orgnaoïdes, expérimentation animale Responsabilité collective : responsable technique de la plate-forme OrgaRES/ORGALille/OrgaNO soumaya.elmoghrabi(@)inserm.fr | |
 | Mouloud SOUIDI, Ingénieur d’étude Université de Lille mouloud.souidi(@)inserm.fr |
 | Dr Fatima LAHDAOUI, PhD, Ingénieur de recherche CDD CPER Cancer |
 | Sonia PAGET, Ingénieur d’étude CDD Inserm Compétences : biologie moléculaire et cellulaire, modèles organoïdes, technologie Crispr-Cas9 sonia.paget(@)inserm.fr |
 | Nicolas STOUP (D4 doctorant) nicolas.stoup(@)inserm.fr |
 | Thomas Meynard (D2 doctorant) thomas.meynard(@)cnrs.fr |
Felix Royer (D2, doctorant) felix.royer.etu(@)univ-lille.fr | |
 | Elsa HADJ-BACHIR (D3, Doctorante) elsa.hadj-bachir(@)inserm.fr |
Our scientific strategy and projects for 2020-2024 are based 1- on the large amount of data acquired during the past 5 years regarding tumor resistance mechanisms and biomarkers in oeso-gastric, pancreatic, colonic and renal cancers (see list of publications) and 2- on the creation of the ONCOLille Cancer Institute, to which belong CANTHER, in 2020 on the Lille hospital campus that will aim at developing basic/translational cancer research on tumor resistance and dormancy.
Our team project focuses on mechanisms responsible for tumor resistance to chemotherapeutic treatments with emphasis on: 1- the role of membrane mucins, 2- the role of epigenetic mechanisms in the plasticity of the cancer cell and 3- the molecular comparison of primary and recurrent tumor (see figure below). This should allow us (i) to bring new knowledge regarding these mechanisms and (ii) to develop new models and find new prognostic/predictive markers to better manage patients and offer better treatment (precision medicine) in collaboration with clinicians, pathologists and biologists of the team.
1- Roles of MUC1 and MUC4 in chemoresistance
Our project aims at understanding the roles of the membrane bound mucins MUC1 and MUC4 in carcinogenesis and chemoresistance. We are using a triple approach (in vitro, in vivo and ex vivo). We especially chose highly resistant cancers (kidney and pancreas) in which MUC1 and MUC4 are overexpressed to better understand their role in these mechanisms (Jonckheere 2017; Jonckheere 2014; Jonckheere 2010; Jonckheere 2008).
1.1. MUC1 (PI: Michael Perrais ; Co-Investigators: Dr Viviane Gnemmi, Pr Sébastien Aubert, Dr Jean-Baptiste Gibier)
In kidney cancer, we have shown that MUC1 is an actor in chemoresistance to conventional and targeted chemotherapies (Aubert 2009, Bouillez 2014, Gnemmi 2014, Gibier 2017). In this project, we will decipher mechanisms involved in this resistance via MUC1-C cleavage by using transcriptomic and proteomic approaches. By using a mass spectrometry approach, we will determine cleavage sites on MUC1-C (collab. Pr I. Fournier, PRISM laboratory, Villeneuve d’Ascq). We will assay the impact of mutated forms on cancer cell properties (migration, invasion, proliferation) and chemoresistance. We will collaborate with Pr N. Lebegue (LilNCog center, Inserm UMR1172, Lille) to develop a strategy to block MUC1-C cleavage. In parallel, we will develop nanobodies also called VHH antibodies against MUC1-C (Hybrigenics services, Paris). Finally, by using preclinical mouse models and Muc1 KO mice, we will study the relationship between MUC1, inflammatory kidney fibrosis and cancer development (collab. Dr C. Cauffiez, Dr N. Pottier & Pr F. Glowacki/Senescence Canther team).
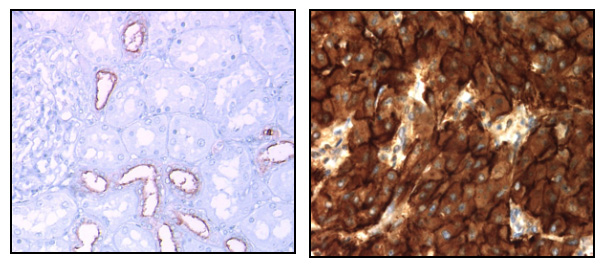 Expression of MUC1 in normal kidney (left) and overexpression in metastatic kidney cancer (right)
Expression of MUC1 in normal kidney (left) and overexpression in metastatic kidney cancer (right)
1.2. MUC4 (PIs: Dr Nicolas Jonckheere, Dr Isabelle Van Seuningen; Co-Investigators: Pr Emmanuelle Leteurtre, Dr Audrey Vincent, Dr Bernadette Neve, Dr Lucie Coppin, Pr Stéphanie Truant, Dr Mehdi El Amrani)
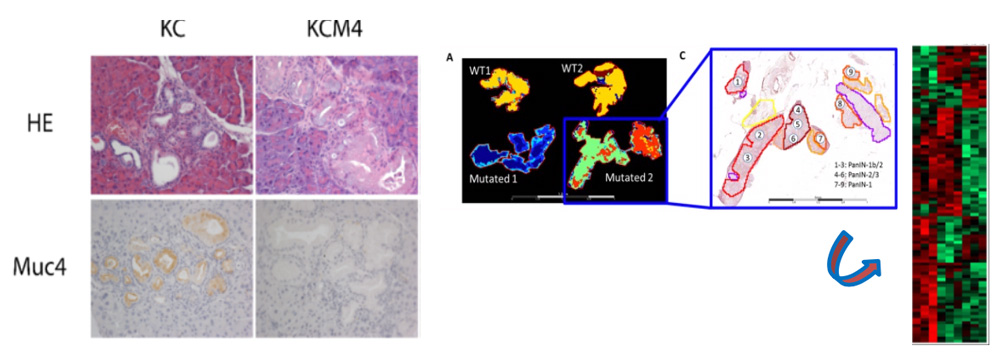 Expression of Muc4 in pancreatic cancer in mice (KC) and in mice deficient for Muc4 (KCM4) (left). Intratumor molecular heterogeneity and PanIN map using MALDI-MSI (right)
Expression of Muc4 in pancreatic cancer in mice (KC) and in mice deficient for Muc4 (KCM4) (left). Intratumor molecular heterogeneity and PanIN map using MALDI-MSI (right)
1.2.a. MUC4 and early pancreatic lesions
Pancreatic adenocarcinoma (PDAC) is a very complex disease harboring intratumoral heterogeneity (ITH) between tumor cells, PanIN lesions (PanIN1a-b/2/3) and highly fibrotic stroma. We aim to elucidate the role of MUC4 in pancreatic carcinogenesis initiation by characterizing the preclinical triple transgenic mouse model KCM4 (Pdx1-Cre; LstopL-KrasG12D; Muc4KO). We want to phenotypically characterize the KCM4 PanINs and establish their cartography and their proteomic signature by MALDI MSI (collab. Pr I. Fournier, PRISM laboratory, Villeneuve d’Ascq). The identified new markers of interest (MOI) will be further studied for their biological roles in PDAC. With pathologists, we aim at finding new biomarkers of response using the retrospective “long survivor” PDAC patient cohort (CAPALONG), we have established in collaboration with hospitals from the inter-region (Hauts de France-Normandie/Cancéropôle Nord-Ouest) (Coordinator: Pr E Leteurtre). With bioinformaticians we will develop new tools using artificial intelligence (machine/deep learning) to identify early lesions and stratify patients and orientate treatments (collab. Pr M. Elati, Plasticity Canther team).
1.2.b. MUC4 and chemoresistance
We aim to decipher the molecular mechanisms responsible for drug resistance (innate and acquired) in PDAC. For that we have established in vitro (2D chemoresistant cell lines), in vivo (chemoresistant patient-derived xenograft (PDX) models), and 3D organoid models. We are also developing innovative “tumor-on-a-chip” 4D microfluidic models in collaboration with UMI2820 CNRS/LIMMS (Dr V. Senez) of the ONCOLille institute. Using loss and gain-of-function strategies (shRNA, Crispr/Cas9), we will investigate the role of MUC4 in resistance to gemcitabine/FOLFIRINOX regimen. MOI identified will be validated for their therapeutic effect in in vivo pre-clinical murine models. We will notably investigate involvement of ion channels (TRPM7, KCa3.1; collab. Pr H. Ouadid-Aidouch, UPJV, Amiens) and miRNAs (miR-210, miR-132-3p, miR-96-5p) associated with MUC4 expression. We will discriminate common molecular patterns that universally drive chemoresistance in PDAC from drug-specific resistance pathways and study the influence of CAFs on tumor behavior (collab. Dr C. Bousquet, CRCT Toulouse, INCa PAIR Pancreas). Finally, the 4D microfluidic device will be used to study: 1- PDAC cell resistance to treatments, 2- Influence of stromal cells in triggering the chemo-induced resistance (EMT, epigenetics) and 3- Increase the throughput of our drug screening on organoids obtained from fine-needle aspirate biopsies (collab. Dr J. Branche, Digestive Diseases Department, Lille Hospital).
1.2.c. MUC4 as a therapeutic target?
The MUC4-ErbB2 complex may represent a critical therapeutic target in PDAC. Having identified the region involved in the interaction (Liberelle 2019, Liberelle 2020), we develop in collaboration with chemists (Pr N. Lebègue, LilNCog, Inserm UMR1172, Lille) a program to find new therapeutic molecules targeting this interaction. The aim of this program is to patent new therapeutic molecules.
2- Epigenetic plasticity of the tumor cell
(PIs: Dr Audrey Vincent, Dr Bernadette Neve; Co-Investigators: Pr Pascal Pigny, Dr Julie Leclerc, Dr Anne-Frédérique Dessein, Dr Florence Renaud, Pr Stéphanie Truant)
Aberrant epigenetic changes are known to ensure cancer cell properties to switch CSC markers on and off, generating a heterogeneous population of cells (Vincent & Van Seuningen 2012). Our recent work suggests that a better knowledge of epigenetic mechanisms that drive cancer stemness could help (i) identify druggable epigenetic modulators, (ii) discriminate tumors that can benefit from “first-generation” epigenetic modifier inhibitors, (iii) design new epidrugs that would be more specific of cancer stemness/plasticity, (iii) revert chemoresistance and (iv) prevent cancer predisposition or relapse (Vincent 2019).
We will use two cancer models, well-known in the laboratory, that take advantage of different stemness properties: colorectal cancer (CRC) through tumor dormancy and relapse, and pancreatic adenocarcinoma (PDAC) through rapid chemoresistance. Using functional stem cell assays including 3D culture (adherent and non-adherent, microfluidics) and xenograft models we will systematically compare CSC to non-CSC at different levels of epigenetic modulation.
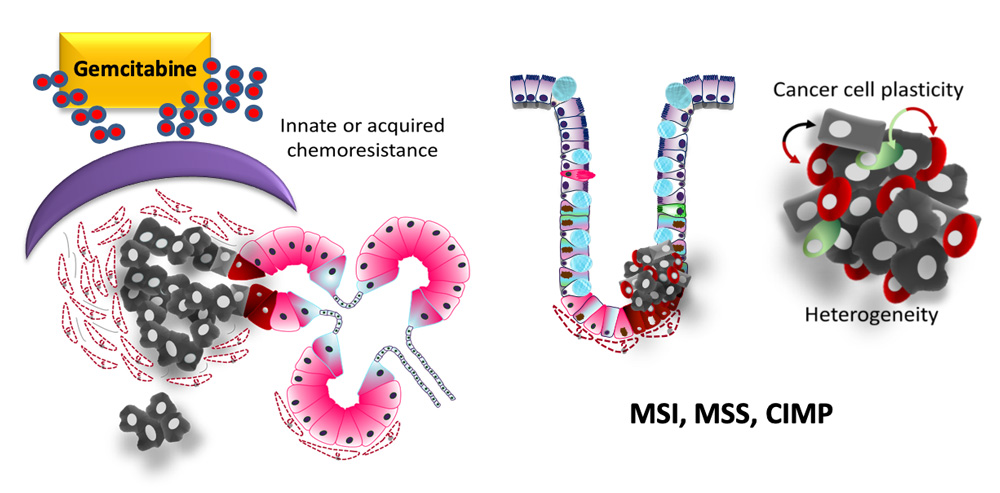 Pancreatic cancer and innate or induced chemoresistance (left). Colonic cancer cell plasticity, dormancy and tumor heterogeneity (right)
Pancreatic cancer and innate or induced chemoresistance (left). Colonic cancer cell plasticity, dormancy and tumor heterogeneity (right)
2.1. Epigenetic modulators
Among direct epigenetic modulators, we aim to identify long non-coding RNAs (lncRNA) that potentially drive cellular plasticity (Neve 2018). We will also study the role of metabolism on chromatin modifications. We will use metabolomics (collab. Dr J.F. Goossens, Mass Spectrometry platform, U. Lille), identify a metabolic signature of cancer cell plasticity (collab. Pr P. Marchetti/Leukemia CANTHER team) and evaluate the role endoplasmic reticulum stress on epigenetic changes (collab. Dr D. Delacour, UMR7592 CNRS, Institut Jacques Monod, Paris).
2.2. Epigenetic modifiers
We aim to study epienzyme and chromatin signatures of normal and CSC using transcriptomics and epigenome-wide approaches in different molecular subtypes of CRC (CIN, MSI, CIMP) and PDAC (epithelial vs basal-like). We will use KD approaches or epigenome edition. We will identify targetable epienzyme-epienzyme interactions for the discovery of stemness-specific new chemical compounds (collab. Pr A-S Voisin, CERMN, Univ. Caen, France).
2.3. Epigenetic mediators
We will continue our work on the molecular mechanisms and the consequences of MLH1 and MUC5AC promoter methylation in familial and sporadic MSI CRC (Leclerc 2018 ; Renaud 2015, 2016). We will use epigenome-editing (CRISPR-dCAs9), vectors enabling tracing of DNA methylation dynamic changes at single-cell resolution and Chromatin Conformation Capture (3C) (collab. Fonctional genomic & Molecular Biology platform (Dr M. Figeac) & BiLille platform (Dr G. Marot), PLBS UMS2014-US41). For that, we have set up the largest diagnostic French cohort of Lynch syndrome patients with constitutional MLH1 promoter methylation (Leclerc 2020 Inserm interface grant). Our long-term goal will be to design new specific epidrugs that would control the methylation state of MLH1 and MUC5AC promoters (collab. Pr A-S Voisin, CERMN, Univ. Caen, France).
3- Oesogastric cancers recurrence molecular profiling for better patient management
(PI: Dr Florence Renaud; Co-Investigators: Pr Marie-Pierre Buisine, Pr Guillaume Piessen, Dr Clarisse Eveno, Dr Isabelle Van Seuningen, Dr Fatima Lahdaoui)
Here we want to get a precise characterization of the molecular landscape of primary tumor, local recurrence or distant metastasis in order to understand (i) the natural history of OG cancers and (ii) mechanisms of resistance to treatments, to develop a simple robust functional test to use in clinic routine, the final objective being to propose more adapted treatments to the patients.
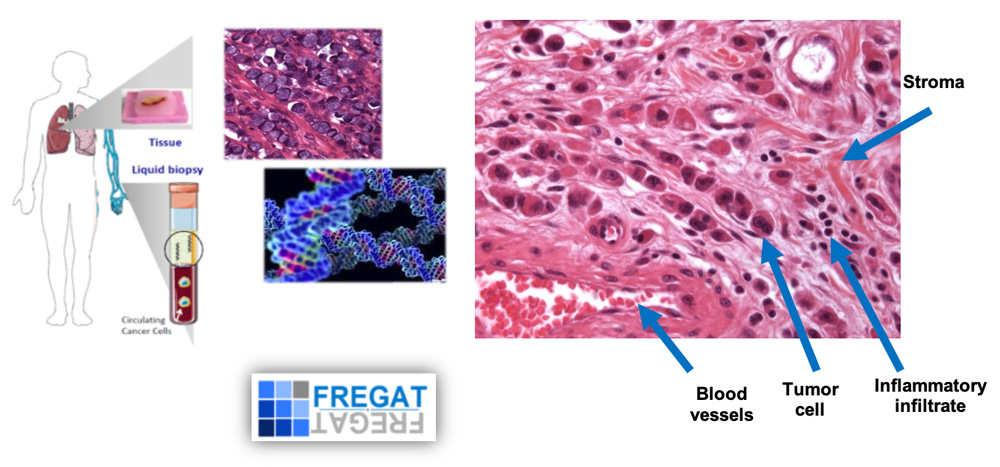 Molecular profiling of the primary tumor and its recurrence in oeso-gastric cancers. Use of the national FREGAT cohort (left). Tumor and cell heterogeneity of an independent cell gastric carcinoma. Tumor-stroma interactions (right).
Molecular profiling of the primary tumor and its recurrence in oeso-gastric cancers. Use of the national FREGAT cohort (left). Tumor and cell heterogeneity of an independent cell gastric carcinoma. Tumor-stroma interactions (right).
3.1. Translational research: Molecular monitoring of recurrence to anticipate resistance of advanced disease
Using our data from PROMOREC and REComics studies, our project aims at evaluating longitudinal monitoring of circulating tumor DNA (ctDNA) using non-invasive liquid biopsy already stored in the FREGAT CBD as a surrogate for tumor recurrence profiling to early detect relapse and to overcome ITH in routine practice. In parallel, using Patient-Derived Organoids (PDOs), we will test effects of drugs targeting the biomarkers identified as new potential targets that could help improve treatments. Moreover, we will determine UPR gene expression signatures associated with response to chemotherapy (collab. Dr O. Pluquet/Senescence CANTHER team).
3.2. Intratumor genetic heterogeneity in gastric PCC as a barrier to precision medicine
Results reported to date strongly suggest that a primary reason for failed targeted therapy trials in metastatic OG cancers is the problem of Intra-Tumor Heterogeneity (ITH). Poorly cohesive carcinoma (PCC) is a particularly aggressive subtype of OG characterized by dramatic increase of incidence, with high level of recurrence and chemoresistance (Piessen 2013, 2009 ; Messager 2011). We will initiate a pilot study with large sampling (cartography) of resected specimen from the FREGAT CBD to characterize molecular landscape of ITH of the primary tumor and synchronous LR localizations (lymph node, peritoneal nodule) both at the DNA, RNA (collab. Pr M.P. Buisine, genomic platforms of the Lille University Hospital & Dr M. Figeac, UMS2014-US41/PLBS) and protein (collab. Pr I. Fournier, PRISM, Villeneuve d’Ascq) levels. This ITH will also be addressed by molecular profiling (DNA) of liquid biopsies. We will also conduct a preliminary study based on an extensive evaluation of stroma reaction on samples before and after chemotherapy. In collaboration with Dr N. Bonnefoy (IRCM, Inserm U1194, Montpellier) and Pr A. Adenis (Medical Oncology Department, IRCM, Montpellier), we will evaluate the tumor immune landscape in response to oxaliplatin-based chemotherapy, an inducer of immune response.
3.3. Development of a tumor-on-chip to investigate drug resistance in gastric PCC
We will create a 4D microfluidic model of tumor-on-chip using droplet-based microfluidics and tissue engineering. This will be carried out in collaboration with the physicists of UMI2820 CNRS/LIMMS laboratory (ONCOLille institute) (Pekin 2011). These models should allow us to better understand the cellular mechanisms involved in drug resistance by isolating the components of the stroma involved in this process and identify or even discover novel cancer biomarkers allowing observation, prediction and elimination of this resistance. This technology should also allow us to develop a new functional clinical test for the routine use in precision medicine (drug resistance, choice of treatment regimen).
3.4. Identify the determinants of tumor resistance and their consequence on patient survival
The determinants of tumor resistance, cancer progression and their consequences for cancer patient survival is a major public health issue (Lelorain et al. 2018). Using the FREGAT CBD, which prospectively collects clinical, biological and psychological data from OG cancer patients at the main stages of their treatments, we will evaluate the association between depressive and/or anxious symptoms with (i) the post-operative morbimortality for OG cancer and (ii) inflammation, immune and tumor markers. The results will provide a better understanding of the role of affective and biological symptoms in post-operative morbimortality and will contribute to its reduction by assessing and treating the affective symptoms at the early stages of treatment (collab. Pr V. Christophe, Dr D. Grynberg, SCALab, ONCOLille institute).
Overall, our team project should provide new fundamental knowledge about the role of mucins in cancer but also on the mechanisms driving tumor cell resistance to the chemotherapeutic drugs, their escape to the drug action, their capacity to metastasize, the role of stroma and on the role of epigenetic modifications and of non-coding RNAs on cancer cell plasticity and capacity to reprogram. Interdisciplinary projects with chemists, physicists, bioinformaticians, will help develop new original models to answer the central question of drug resistance. Translational programs with clinicians and pathologists of the team, using patient cohorts and CBD data and collaboration with HSS colleagues, should help better define molecularly sub-types of cancers and find new prognostic/predictive/response markers to provide better management and patient care.
> ORIGINAL ARTICLES FROM THE LABORATORY
> GENERAL REVIEWS
> EDITORIAL
> LETTERS – COMMENTARIES
> BOOK CHAPTERS
> ARTICLES AND REVIEWS FROM COLLABORATIONS
> CLINICAL PUBLICATIONS
2021
Gibier JB*, Swierczewski T*, Csanyi M, Hémon B, Glowacki FX, Maboudou P, Van Seuningen I, Cauffiez C, Pottier N, Aubert N, Perrais M, Gnemmi V. MUC1 mitigates renal injury and inflammation in endotoxin induced acute kidney injury by inhibiting the TLR4-MD2 axis and reducing pro-inflammatory macrophages infiltration. Shock 2021 Jan 28. doi: 10.1097/SHK.0000000000001742.
Bauzone* M, Souidi M*, Dessein AF, Wisztorski M, Vincent A, Gimeno JP, Monté D, Van Seuningen I, Gespach C, Huet G. Cross-talk between YAP and RAR-RXR Drives Expression of Stemness Genes to Promote 5-FU Resistance and Self-Renewal in Colorectal Cancer Cells. Mol Cancer Res. 2021 Jan 20. doi: 10.1158/1541-7786.MCR-20-0462. IF: 4.630
2020
Jonckheere N, Auwercx J, Hadj Bachir E, Coppin L, Boukrout N, Vincent A, Neve B, Gautier M, Treviño V, Van Seuningen I. Unsupervised hierarchical clustering of pancreatic adenocarcinoma dataset from TCGA defines a mucin expression profile that impacts overall survival. Cancers (Basel). 2020 Nov 9;12(11):3309. doi: 10.3390/cancers12113309. IF: 6.126
Coppin L, Jannin A, Ait Yahya E, Thuillier C, Villenet C, Tardivel M, Bongiovanni A, Gaston C, de Beco S, Barois N, van Seuningen I, Durand E, Bonnefond A, Vienne JC, Vamecq J, Figeac M, Vincent A, Delacour D, Porchet N, Pigny P. Galectin-3 modulates epithelial cell adaptation to stress at the ER-mitochondria interface. Cell Death Dis. 2020 May 12;11(5):360. IF: 6.304
Roberti MP, Yonekura S, Duong CPM, Picard M, Ferrere G, Tidjani Alou M, Rauber C, Iebba V, Lehmann CHK, Amon L, Dudziak D, Derosa L, Routy B, Flament C, Richard C, Daillère R, Fluckiger A, Van Seuningen I, Chamaillard M, Vincent A, Kourula S, Opolon P, Ly P, Pizzato E, Becharef S, Paillet J, Klein C, Marliot F, Pietrantonio F, Benoist S, Scoazec JY, Dartigues P, Hollebecque A, Malka D, Pagès F, Galon J, Gomperts Boneca I, Lepage P, Ryffel B, Raoult D, Eggermont A, Vanden Berghe T, Ghiringhelli F, Vandenabeele P, Kroemer G, Zitvogel L. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nature Medicine, 2020 Jun;26(6):919-931. doi: 10.1038/s41591-020-0882-8. Epub 2020 May 25. PMID: 3245149.
2019
Liberelle M, Magnez R, Thuru X, Bencheikh Y, Ravez S, Quenon C, Drucbert AS, Foulon C, Melnyk P, Van Seuningen I*, Lebègue N*. MUC4-ErbB2 Oncogenic Complex: Binding studies using Microscale Thermophoresis and Surface Plasmon Resonance. Sci Rep, 2019 Nov 13;9(1):16678. doi: 10.1038/s41598-019-53099-0
El Amrani M, Corfiotti F, Corvaisier M, Vasseur R, Fulbert M, Skrzypczyk C, Deshorgues AC, Gnemmi V, Tulasne D, Lahdaoui F, Vincent A, Pruvot FR, Van Seuningen I, Huet G, Truant S. Gemcitabine-induced epithelial-mesenchymal transition-like changes sustain chemoresistance of pancreatic cancer cells of mesenchymal-like phenotype. Mol Carcinog. 2019 Aug 2. doi: 10.1002/mc.23090.
2018
Jonckheere N, Van Seuningen I. Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20 signature is associated with poor survival in human carcinomas. J Transl Med. 2018 Sep 20;16(1):259. doi: 10.1186/s12967-018-1632-2.
Drubay V, Skrypek N, Cordiez L, Vasseur R, Schulz C, Boukrout N, Duchêne B, Coppin L, Van Seuningen I, Jonckheere N. TGF-βRII Knock-down in Pancreatic Cancer Cells Promotes Tumor Growth and Gemcitabine Resistance. Importance of STAT3 Phosphorylation on S727. Cancers (Basel). 2018 Jul 31;10(8). pii: E254. doi: 10.3390/cancers10080254.
2017
Lahdaoui F, Messager M, Vincent A, Hec F, Gandon A, Warlaumont M, Renaud F, Leteurtre E, Piessen G, Jonckheere N, Mariette C, Van Seuningen I. Depletion of MUC5B mucin in gastrointestinal cancer cells alters their tumorigenic properties: Implication of the Wnt/β-catenin pathway. 2017 Biochem J. 2017 Nov 1;474(22):3733-3746.
Gaudelot K, Gibier J-B, Pottier N, Hémon B, Van Seuningen I, Glowacki F, Leroy X, Cauffiez C, Gnemmi V, Aubert S, Perrais M. Anti-miR-21 strategy promotes chemosensitivity and decreases expression of multi-drug resistant genes in renal carcinoma. Tumor Biol, 2017 Jul;39(7):1010428317707372. doi: 10.1177/1010428317707372.
Coppin L, Vincent A, Frénois F, Duchene B, Lahdaoui F, Stechly L, Renaud F, Villenet C, Van Seuningen I, Leteurtre E, Dion J, Grandjean C, Poirier F, Figeac M, Delacour D, Porchet N, Pigny P. Galectin-3 is a non-classic RNA binding protein that stabilizes the mucin MUC4 mRNA in the cytoplasm of cancer cells. Sci Rep. Mar 6;7:43927.
Gibier, J-B, Hémon B, Fanchon M, Gaudelot K, Pottier N, Ringot B, Van Seuningen I, Glowacki F, Cauffiez C, Blum D, Copin M-C, Perrais M, Gnemmi V. Dual role of MUC1 mucin in kidney ischemia-reperfusion injury: nephroprotector in early phase, but pro-fibrotic in late phase. Biochim Biophys Acta-Mol Basis Dis, 2017 Mar 31;1863(6):1336-1349.
2016
Corvaisier M, Bauzone M, Corfiotti F, Renaud F, El Amrani M, Monté D, Truant S, Leteurtre E, Formstecher P, Van Seuningen I, Gespach C, Huet G. Regulation of cellular quiescence by YAP/TAZ and cyclin E1 in colon cancer cells: implication in chemoresistance and cancer relapse. Oncotarget, 2016 Aug 30;7(35):56699-56712.
Coppin L, Benomar K, Corfiotti F, et al. CA-125, but not galectin-3, complements CA 19-9 for discriminating ductal adenocarcinoma versus non-malignant pancreatic diseases. Pancreatology 2016; 16: 115-120.
Renaud F, Mariette C, Vincent A, Wacrenier A, Maunoury V, Leclerc J, Coppin L, Crépin M, Van Seuningen I, Leteurtre E, Buisine MP. The serrated neoplasia pathway of colorectal tumors: Identification of MUC5AC hypomethylation as an early marker of polyps with malignant potential. Int J Cancer. 2016 Mar 15;138(6):1472-81.
2021
Auwercx J, Rybarczyk P, Kischel P, Dhennin-Duthille I, Chatelain D, Sevestre H, van Seuningen I, Ouadid-Ahidouch H, Jonckheere N, Gautier M. Mg2+ transporters in digestive cancers. Nutrients. 2021 Jan 13;13(1):210.
2020
Neve B, Jonckheere N, Vincent A, Van Seuningen I. Long non-coding RNAs: the tentacles of chromatin remodeler complexes. Cell Mol Life Sci. 2020 Oct 1. doi: 10.1007/s00018-020-03646-0.
Liberelle M, Jonckheere N, Melnyk P, Van Seuningen I, Lebegue N. EGF-containing Membrane-bound Mucins: a hidden ErbB2 targeting pathway? J Med Chem. 2020 May 28;63(10):5074-5088
2019
Vincent A, Ouelkdite-Oumouchal A, Souidi M, Leclerc J, Neve B, Van Seuningen I. Colon cancer stemness as a reversible epigenetic state: Implications for anticancer therapies. World J Stem Cells, 2019 Nov 26:11(11):920-936.
2018
Neve B, Jonckheere N, Vincent A, Van Seuningen I. Epigenetic Regulation by lncRNAs: An Overview Focused on UCA1 in Colorectal Cancer. Cancers (Basel). 2018 Nov 14;10(11). pii: E440. doi: 10.3390/cancers10110440.
Coppin L, Leclerc J, Vincent A, Porchet N, Pigny P. Messenger RNA life-cycle in cancer cells: Emerging role of conventional and non-conventional RNA-binding proteins? Int J Mol Sci, 2018, 19, 650; doi:10.3390/ijms19030650.
2017
Jonckheere N, Vasseur R, and Van Seuningen I. The cornerstone K-ras mutation in pancreatic adenocarcinoma: from cell signaling network, target genes, biological processes to therapeutic targeting. Crit Rev Oncol Hematol, 2017 111: 7-19.
2019
Jonckheere N, Van Seuningen I. Fine-tuning autophagy in pancreatic adenocarcinoma: full blockage is required. Ann Transl Med. 2019 Mar;7(Suppl 1):S43.
2020
Neve B, Jonckheere N, Vincent A, Van Seuningen, I. 2020. Single cell analysis may shed new lights on the role of lncRNAs in chemo-resistance in gastrointestinal cancers. In: RNA technology. Springer Series. The chemical biology of long non coding RNAs. Volume 10. Ed. S. Jurga & J. Barciszewski.
ARTICLES AND REVIEWS FROM COLLABORATIONS
2020
Roberti MP, Ferrere G, Tidjani Alou M, Duong CMP, Routy B, Becharef S, Goéré D, Flament C, Poirier-Colame V, Richard C, Daillère R, Flückiger A, Van Seuningen I, Chamaillard M, Vincent A, Merad M, Enot D, Stoll G, Kourula S, Vétizou M, Yamazaki T, Fahrner JE, Arnould L, Ladoire S, Guyard L, Marliot F, Paci A, Broutin S, Bonnet R, Sauvanet P, Pezet D, Benoist S, Scoazec JY, Malka D, Pagès F, Galon J, Gomperts Boneca I, Lepage P, Raoult D, Eeggermont A, Ghiringhelli F, Vandenabeele P, Kroemer G, Zitvogel L. The Ileal Microbiome Shapes the Immunosurveillance and Prognosis of Proximal Colon Cancer. Nature Medicine, sous presse/in press
2019
Lambert M, Alioui M, Jambon S, Depauw S, Van Seuningen I, David-Cordonnier M-H. Direct and indirect targeting of HOXA9 transcription factor in acute myeloid leukemia. Cancers. 2019 Jun 17;11(6). IF: 5.326
2018
Vandenbussche C, Van der Hauwaert C, Dewaeles E, Franczak J, Hennino MF, Gnemmi V, Savary G, Tavernier Q, Nottet N, Paquet A, Perrais M, Blum D, Mari B, Pottier N, Glowacki F, Cauffiez C. Tacrolimus-induced nephrotoxicity in mice is associated with microRNA deregulation. Arch Toxicol. 2018 Jan 23. doi: 10.1007/s00204-018-2158-3
Aubert S, Berdelou A, Gnemmi V, Behal H, Caiazzo R, D’herbomez M, Pigny P, Wemeau JL, Carnaille B, Renaud F, Bouchindhomme B, Leteurtre E, Perrais M, Pattou F, Do Cao C. Large sporadic thyroid medullary carcinomas: predictive factors for lymph node involvement. Virchows Arch. 2018 Feb 1. doi: 10.1007/s00428-018-2303-7.
2017
Gibault F, Bailly F, Corvaisier M, Coevoet M, Huet G, Melnyk P, Cotelle P. Molecular features of the YAP inhibitor, Verteporfin ‐ synthesis of hexasubstituted dipyrrins as potential inhibitors of YAP/TAZ, the downstream effectors of the Hippo pathway. (2017) Chem Med Chem. 12, 954-961
Duruisseaux M, Antoine M, Rabbe N, Rodenas A, Mc Leer-Florin A, Lacave R, Poulot V, Duchêne B, Van Seuningen I, Cadranel J, Wislez M. Lepidic predominant adenocarcinoma and invasive mucinous adenocarcinoma of the lung exhibit specific mucin expression in relation with oncogenic drivers. Lung Cancer. 2017 Jul;109:92-100.
De Arcangelis A, Hamade H, Alpy F, Normand S, Bruyère E, Lefebvre O, Méchine-Neuville A, Siebert S, Pfister V, Lepage P, Laquerriere P, Dembele D, Delanoye-Crespin A, Rodius S, Robine S, Kedinger M, Van Seuningen I, Simon-Assmann P, Chamaillard M, Labouesse M, Georges-Labouesse E. Hemidesmosome integrity protects the colon against colitis and colorectal cancer. Gut, 2017 Oct;66(10):1748-1760.
Porte R, Van Maele L, Muñoz-Wolf N, Foligné B, Dumoutier L, Tabareau J, Cayet D, Gosset P, Jonckheere N, Van Seuningen I, Chabalgoity JA, Simonet M, Lamkanfi M, Renauld JC, Sirard JC, Carnoy C. Flagellin-mediated protection against intestinal Yersinia pseudotuberculosis infection does not require interleukin-22. Infect Immun. 2017 Jan 26;85(2). pii: e00806-16.
Colomb, F., Krzewinski-Recchi, M.A., Steenackers, A., Vincent, A., Harduin-Lepers, A., Delannoy, P., Groux-Degroote, S. TNF up-regulates ST3GAL4 and sialyl-Lewisx expression in lung epithelial cells through an intronic ATF2-responsive element. Biochem J. 2017 Jan 1;474(1):65-78.
2016
Omura, N., Mizuma, M., MacGregor, A., Hong, S.M., Ayars, M., Borges, M., Kanda, M., Li, A., Vincent, A., Maitra, A., Goggins, M. Overexpression of ankyrin1 promotes pancreatic cancer cell growth. Oncotarget. 2016 Jun 7;7(23):34977-87.
Gibault F, Corvaisier M, Bailly F, Huet, G, Melnyk P, Cotelle P. Non-photoinduced biological properties of verteporfin (2016) Curr. Med. Chem., 23, 1171-1184. (review)
2019
Eveno C, Voron T, Piessen G. Laparoscopic Hyperthermic Intraperitoneal Chemotherapy is Safe for Patients with Peritoneal Metastases from Gastric Cancer and May Lead to Gastrectomy. Ann Surg Oncol. 2019 Jul 8. doi: 10.1245/s10434-019-07603-5.
Mariette C, Markar SR, Dabakuyo-Yonli TS, Meunier B, Pezet D, Collet D, D’Journo XB, Brigand C, Perniceni T, Carrère N, Mabrut JY, Msika S, Peschaud F, Prudhomme M, Bonnetain F, Piessen G; Fédération de Recherche en Chirurgie (FRENCH) and French Eso-Gastric Tumors (FREGAT) Working Group. Hybrid Minimally Invasive Esophagectomy for Esophageal Cancer. N Engl J Med. 2019 Jan 10;380(2):152-162. doi: 10.1056/NEJMoa1805101.
Renaud F, Chetboun M, Thevenet J, Delalleau N, Gmyr V, Hubert T, Bonner C, Messager M, Leteurtre E, Mariette C, Kerr-Conte J, Piessen G, Pattou F. Safety of Islet Autotransplantation After Pancreatectomy for Adenocarcinoma. Transplantation. 2019 Jan;103(1):177-181. doi: 10.1097/TP.0000000000002419.
2018
Mariette C, Renaud F, Piessen G, Gele P, Copin MC, Leteurtre E, Delaeter C, Dib M, Clisant S, Harter V, Bonnetain F, Duhamel A, Christophe V, Adenis A, on behalf of the FREGAT working group. The FREGAT biobank: a clinico-biological database dedicated to esophageal and gastric cancers. BMC Cancer 2018 Feb 6;18(1):139
2017
Mariette C. What is the optimal neoadjuvant treatment for locally advanced oesophageal adenocarcinoma? Ann Oncol. 2017 Mar 1;28(3):447-450.
Renaud F, Bibeau F, Leteurtre E, Delaeter C, Dib M, Harter V, Adenis A, Piessen G, Mariette C. Base clinico-biologique nationale française dédiée à la recherche sur les cancers oeso-gastriques FREGAT: French clinicobiological database dedicated to esogastric cancers. Ann Pathol. 2017 Dec;37(6):457-466.
Mantziari S, Gronnier C, Renaud F, Duhamel A, Théreaux J, Brigand C, Carrère N, Lefevre JH, Pasquer A, Demartines N, Collet D, Meunier B, Mariette C; FREGAT working group – FRENCH – AFC. Survival Benefit of Neoadjuvant Treatment in Clinical T3N0M0 Esophageal Cancer: Results From a Retrospective Multicenter European Study. Ann Surg. 2017 Nov;266(5):805-813.
Degisors S, Pasquer A, Renaud F, Béhal H, Hec F, Gandon A, Vanderbeken M, Caranhac G, Duhamel A, Piessen G, Mariette C; FREGAT working group. Are Thoracotomy and/or Intrathoracic Anastomosis Still Predictors of Postoperative Mortality After Esophageal Cancer Surgery?: A Nationwide Study. Ann Surg. 2017 Nov;266(5):854-862.
Markar SR, Gronnier C, Pasquer A, Duhamel A, Behal H, Théreaux J, Gagnière J, Lebreton G, Brigand C, Meunier B, Collet D, Mariette C; FREGAT working group – FRENCH – AFC. Surgically treated oesophageal cancer developed in a radiated field: Impact on peri-operative and long-term outcomes. Eur J Cancer. 2017 Apr;75:179-189.
M Messager, M Warlaumont, F Renaud, H Marin, J Branche, G Piessen, C Mariette. Recent improvements in the management of esophageal anastomotic leak after surgery for cancer. Eur J Surg Oncol 2017;43:258-269.
2016
Lordick F, Mariette C, Haustermans K, Obermannová R, Arnold D; ESMO Guidelines Committee. Oesophageal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016 Sep;27(suppl 5):v50-v57.
Pasquer A, Renaud F, Hec F, Gandon A, Vanderbeken M, Drubay V, Caranhac G, Piessen G, Mariette C; FREGAT Working GroupFRENCH. Is Centralization Needed for Esophageal and Gastric Cancer Patients With Low Operative Risk?: A Nationwide Study. Ann Surg. 2016 Nov;264(5):823-830.
Voron T, Messager M, Duhamel A, Lefevre JH, Mabrut JY, Goere D, Meunier B, Brigand C, Hamy A, Glehen O, Mariette C, Paye F; On behalf of the FREGAT working group – FRENCH. Is signet-ring cell carcinoma a specific entity among gastric cancers? Gastric Cancer. 2016 Oct;19(4):1027-40.
Mantziari S, Gronnier C, Pasquer A, Gagnière J, Théreaux J, Demartines N, Schäfer M, Mariette C; FREGAT Working Group–FRENCH–AFC. Incidence and Risk Factors Related to Symptomatic Venous Thromboembolic Events After Esophagectomy for Cancer. Ann Thorac Surg. 2016 Sep;102(3):979-84.
Bekkar S, Gronnier C, Renaud F, Duhamel A, Pasquer A, Théreaux J, Gagnière J, Meunier B, Collet D, Mariette C; French Eso-Gastric Tumors (FREGAT) working group, Fédération de Recherche EN CHirurgie (FRENCH) and Association Française de Chirurgie (AFC). Multicentre study of neoadjuvant chemotherapy for stage I and II oesophageal cancer. Br J Surg. 2016 Jun;103(7):855-62.
A Gandon, C Gronnier, F Renaud, P Borde, M Vanderbeken, F Hec, G Piessen, A Adenis, X Mirabel, C Mariette. Esophageal adenocarcinoma: Impact of a large hiatal hernia on outcomes after surgery. Ann Surg 2016;264:862-870.
Markar SR, Gronnier C, Duhamel A, Pasquer A, Théreaux J, Chalret du Rieu M, Lefevre JH, Turner K, Luc G, Mariette C; FREGAT Working Group-FRENCH-AFC. Significance of Microscopically Incomplete Resection Margin After Esophagectomy for Esophageal Cancer. Ann Surg. 2016 Apr;263(4):712-8.
Markar SR, Gronnier C, Pasquer A, Duhamel A, Beal H, Théreaux J, Gagnière J, Lebreton G, Brigand C, Meunier B, Collet D, Mariette C; French Eso-Gastric Tumors (FREGAT) working group – Federation de Recherche EN CHirurgie (FRENCH) – Association Francaise de Chirurgie (AFC). Role of neoadjuvant treatment in clinical T2N0M0 oesophageal cancer: results from a retrospective multi-center European study. Eur J Cancer. 2016 Mar;56:59-68.
Robb WB, Messager M, Dahan L, Mornex F, Maillard E, D’Journo XB, Triboulet JP, Bedenne L, Seitz JF, Mariette C; Fédération Francophone de Cancérologie Digestive, Société Française de Radiothérapie Oncologique, Union des Centres de Lutte Contre le Cancer, Groupe Coopérateur Multidisciplinaire en Oncologie and the French EsoGAstric Tumour working group – Fédération de Recherche En Chirurgie. Patterns of recurrence in early-stage oesophageal cancer after chemoradiotherapy and surgery compared with surgery alone. Br J Surg. 2016 Jan;103(1):117-25.
On-Going Theses
- HADJ-BACHIR Elsa (D1 en 2020 – Thesis director : Audrey VINCENT)
- STOUP Nicolas (D2 en 2020 – Thesis director : Isabelle VAN SEUNINGEN, Nicolas LEBEGUE)
- XX (D2 en 2020 – Thesis director : Florence RENAUD, Isabelle FOURNIER)
- BOUKROUT Nihad (D4 en 2020 – Thesis director : Nicolas JONCKHEERE)
Defended Theses
- DE MIOLLIS Frédérick (28 janvier 2021) directed by Isabelle VAN SEUNINGEN and Vincent SENEZ
- SWIERCZEWSKI Thomas (26 janvier 2021) directed by Michael PERRAIS
- GIBIER Jean-Baptiste (25 janvier 2021) directed by Sébastien AUBERT and Viviane GNEMMI
- EL AMRANI Mehdi (11/12/2019) directed by Isabelle VAN SEUNINGEN and Guillemette HUET
- SAAD Chadi (26/09/18) directed by Marie-Pierre BUISINE and Hélène TOUZET
- GAUDELOT Kelly (23/06/2017) directed by Michael PERRAIS
- COPPIN Lucie (12/06/2017) directed by Pascal PIGNY
- CORVAISIER Matthieu (29/11/2016) directed by Guillemette HUET
Defended HDR
- GNEMMI Viviane (05/03/2020) directed by Isabelle VAN SEUNINGEN